Optimizing the Plasmid Production Process | Role of Analytics
24Apr2024
As the market for gene therapies continues to grow, the need for high quality plasmid (pDNA) is increasing. Many companies are looking to insource pDNA production and purification – usually as a key starting raw material for subsequent viral vector and mRNA production & purification. Many are familiar with lab-scale plasmid isolation via spin-column kits. However, knowledge gained from lab kits is difficult to implement in a large-scale pDNA purification process. pDNA process development comes with a particular set of optimization, scale-up and cGMP implementation challenges. One of the chief concerns is usually the removal of host cell DNA and RNA while maintaining control over shear forces that would damage the plasmid DNA and create the undesirable OC pDNA isoform
The good news is that the lack of experience can be replaced by analytical insight. What once took months to optimize and years to perfect, can be achieved on far shorter timelines with the appropriate analytical tools. To help achieve such short development timelines, in-process analytics should have the capacity to:
- Easily analyse complex, pre-capture process samples
- Separation before quantification – leads to accurate mass balance determination
1. Easy Analysis of Complex, Pre-Capture Process Samples
Even when using a complex sample like E. coli cell lysate, it is easy to determine the initial yield and pDNA quality with the PATfix pDNA analytical platform. This is an important insight that is hard to obtain using other analytical approaches. It can serve as a control step to evaluate the quality of the starting material while providing an accurate quantification of starting material pDNA mass, that informs subsequent recovery/mass balance calculations.
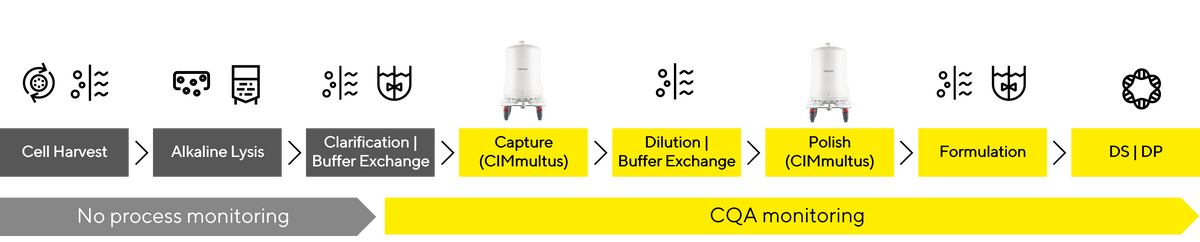
Once the quality of the starting material is established, the next key step is lysis and neutralization of the E. coli resuspension. Optimizing this step represents a significant challenge, when transitioning from lab-scale to pilot scale. Two main considerations that will impact the quality of product after scale-up:
- Precise control over the duration of alkaline lysis, especially when mixing larger volumes. Inconsistencies in this step can lead to yield loss
- Effective control over RNA and cell debris precipitation using calcium chloride influences how easy and repeatable the downstream processing will be.
When lysis is not properly executed, the following problems can occur:
- Increase of open circular (OC) pDNA due to uncontrolled lysis
- Increase of genomic DNA fragments due to uncontrolled lysis
PATfix analytical system offers a guided way to determine RNA and SC/OC pDNA content in these challenging samples, without the need for complex sample pre-treatment. This allows for appropriate & early process insight, and leads to better, repeatable process outcomes.
2. Separation Before Quantification – A Single Analytical Method for Accurate Mass Balance Determination
In a typical cell lysate, the desired plasmid pDNA represents only about 3% of the total content. Equally, genomic DNA (gDNA) also accounts for 3%, and RNA is present at a higher proportion of 21%. Both genomic DNA and RNA are undesirable process contaminants that can also interfere with analytical measurements. All nucleic acids contribute to UV absorbance at 260 nm, a common detection method in molecular biology, while specific dyes are known to have off-target binding. This makes accurate quantification difficult.
E. coli Lysate Composition With Corresponding Analytical Chromatogram

The challenge of accurate quantification is compounded by the fact that the absorbance of nucleic acids is not fixed. It varies with changes in sample properties and is particularly sensitive to alterations in salt concentration and pH levels. This can affect analytics measurements from various stages of the process, such as chromatography fractions (load, flow-through, wash, elution or cleaning-in-place) and as a consequence recovery/mass balance calculation can be skewed.
PATfix analytical chromatography offers a straightforward solution to these issues. Chromatographic separation of key components before their quantification eliminates the problem of interference – impurities are separated from pDNA before UV absorbance measurement. Additionally, because each component consistently elutes at the same retention time in the analytical chromatogram, they do so under identical buffer conditions, regardless of the initial composition of the solution. Analytical chromatography effectively prevents the absorbance measurement being skewed due to initial sample conditions, making it much more reliable.
Taken together, utilizing a reliable analytical method, that can be utilized on all samples in the pDNA purification process – from E. coli cell paste to purified supercoiled pDNA – gives a holistic picture of your process. This allows for much easier optimization of process parameters to achieve target pDNA recoveries and quality.
Convinced of the merits of analytical chromatography, one might wonder, "Why don’t I develop this analytical method on my own?"
This is not necessary, because you can adopt a pre-validated, thoroughly tested system that we've crafted—the PATfix pDNA platform. Get a complete picture of your process by having the ability to reliably analyse all process samples from E.coli lysate to purified SC pDNA, while monitoring CQAs of RNA removal, OC pDNA creation/removal and SC pDNA concentration.
